In the Computational Neurology group, we answer clinical relevant questions in the field of neuropsychiatry using computational methods. We analyze clinical and neuroimaging data using both a data-driven and a modelling-based approach (see Figure 1). This integration helps generating a better (qualitative) and robust (quantitative) understanding of pathophysiological processes of neurological diseases, as well as their diagnosis and treatment.
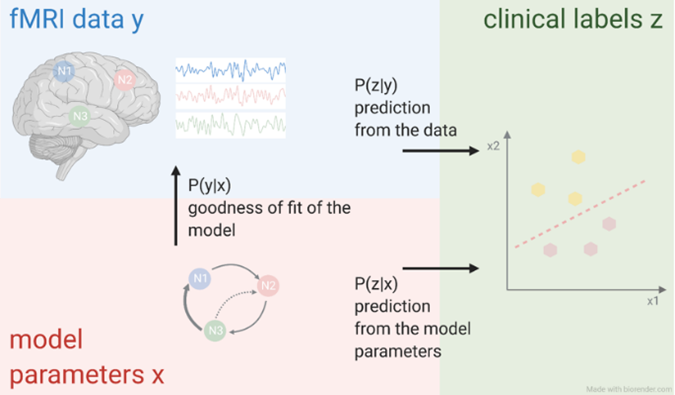
Figure 1: Computational neurology connects a data-driven approach (top left), a model-based approach (bottom left), and clinical data (right). © Xenia Kobeleva
The data-driven approach mainly focusses on the analysis of neurological diseases with methods of machine learning in order to identify characteristic properties of the brain structure and brain function as clinical phenotypes. The model-driven approach builds on our findings from the data-driven approach. It formalizes a mechanistic relationship between clinical data and brain function via equations; in doing so, this relationship can be meaningfully interpreted, and it extends and goes beyond mere observations. Simulated brain models exert high congruence with actual brain data (compare Figure 2).
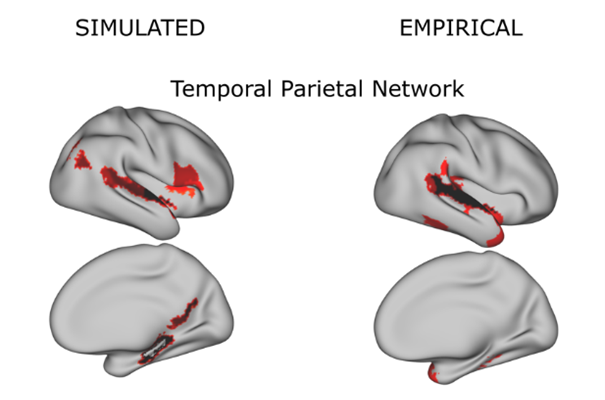
Figure 2: Brain networks can be realistically simulated with dynamical models. The simulated brain network (left) shows high conformity to the actual clinical data (right). © Xenia Kobeleva
This kind of modelling might enable medical doctors to adapt their treatments based on patient-specific mathematical models.
Methods we use in the research group:
- Dynamical modelling (dynamic mean field, Hopf, etc.)
- Analyses of connectivity (structural connectivity, functional connectivity, dynamical functional connectivity, effective connectivity)
- Pre-processing of structural and functional MRT data (including preprocessing pipelines)
- Analyses of resting state fMRT and task fMRT
- Neurophysiology (TMS, EMG, EEG)
- Medical informatics
- Open science



